The Python API is a script that allows access to your MOLGENIS data from python. It is available on every MOLGENIS with version 1.14.0 and up on the URL http://molgenis.mydomain.example/molgenis.py.
Example
As an example, let's create a plot for publicly available ASE data available on https://molgenis56.target.rug.nl/. For a description of the data, take a look at http://molgenis.org/ase.
We'll be creating a scatter plot so if you haven't already, install matplotlib from the commandline:
pip install matplotlib
Download the python api from a molgenis server, for instance https://molgenis01.target.rug.nl/molgenis.py and save it in molgenis.py.
Start an interactive python shell and create a molgenis connection:
import molgenis
This imports the molgenis package.
session = molgenis.Session("https://molgenis56.target.rug.nl/api/")
Instantiates a new Session pointing at the molgenis56 server. If you take a look at the connection by typing
dir(session)
you should see, amongst others, the methods you can call:
[...,'add', 'add_all', 'delete', 'get', 'get_attribute_meta_data', 'get_entity_meta_data', 'login', 'logout',...]
Let's load some data from the server using session.get:
session.get("ASE")
This retrieves the top 1000 rows from the ASE entity.
[{u'Alternative_allele': u'A', u'P_Value': 2.06504739339637e-17, u'Genes': {u'href': u'/api/v1/ASE/rs9901673/Genes'}, u'Fraction_alternative_allele': 0.479, u'Pos': 7484101, u'Reference_allele': u'C', u'Chr': u'17', u'href': u'/api/v1/ASE/rs9901673', u'Samples': u'145', u'Likelihood_ratio_test_D': 72.0813644150712, u'SNP_ID': u'rs9901673'}, {u'Alternative_allele': u'T', u'P_Value': 8.78109735398113e-18, u'Genes': {u'href': u'/api/v1/ASE/rs2597775/Genes'}, u'Fraction_alternative_allele': 0.479, u'Pos': 17503382, u'Reference_allele': u'C', u'Chr': u'4', u'href': u'/api/v1/ASE/rs2597775', u'Samples': u'359', u'Likelihood_ratio_test_D': 73.769089117417, u'SNP_ID': u'rs2597775'}, {u'Alternative_allele': u'C', u'P_Value': 1.4917458949834e-18, u'Genes': {u'href': u'/api/v1/ASE/rs3216/Genes'}, u'Fraction_alternative_allele': 0.479, u'Pos': 214421, u'Reference_allele': u'G', u'Chr': u'11', u'href': u'/api/v1/ASE/rs3216', u'Samples': u'301', u'Likelihood_ratio_test_D': 77.2691957930797, u'SNP_ID': u'rs3216'}, [...],{u'Alternative_allele': u'T', u'P_Value': 0.000132500824069775, u'Genes': {u'href': u'/api/v1/ASE/rs1056019/Genes'}, u'Fraction_alternative_allele': 0.482, u'Pos': 41337435, u'Reference_allele': u'C', u'Chr': u'12', u'href': u'/api/v1/ASE/rs1056019', u'Samples': u'47', u'Likelihood_ratio_test_D': 14.605874945467, u'SNP_ID': u'rs1056019'}]
Let's retrieve a specific SNP from the ASE entity:
print(session.get("ASE", q=[{"field":"SNP_ID", "operator":"EQUALS", "value":"rs12460890"}]))
[{u'Alternative_allele': u'T', u'P_Value': 7.1708540619282e-14, u'Genes': {u'href': u'/api/v1/ASE/rs12460890/Genes'}, u'Fraction_alternative_allele': 0.527, u'Pos': 829568, u'Reference_allele': u'C', u'Chr': u'19', u'href': u'/api/v1/ASE/rs12460890', u'Samples': u'21', u'Likelihood_ratio_test_D': 56.0207947348388, u'SNP_ID': u'rs12460890'}]
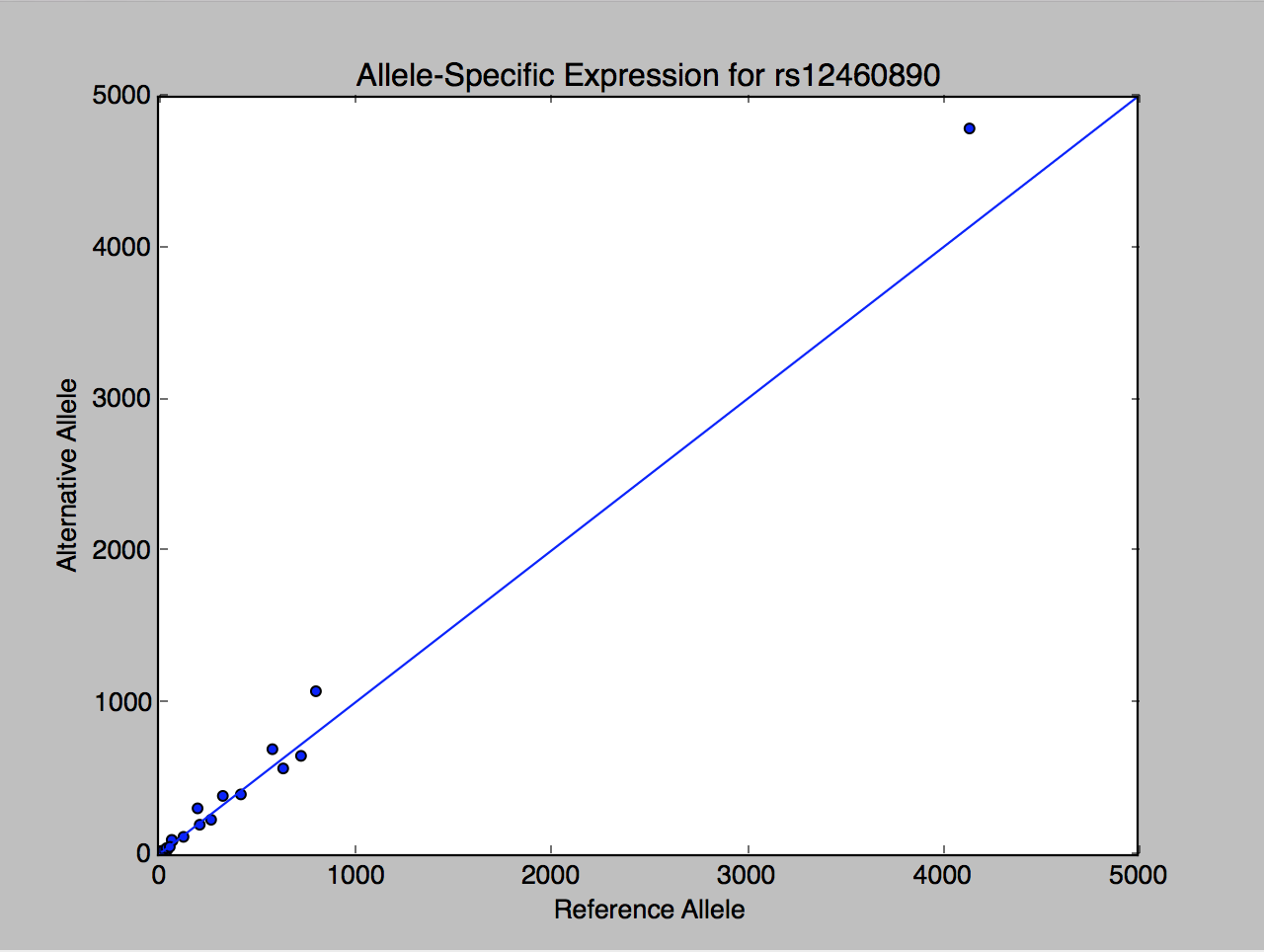
This SNP has a mild but significant allele-specific expression, based on expression counts in 21 samples.
Let's retrieve the samples for this SNP:
samples = session.get("SampleAse", q=[{"field":"SNP_ID", "operator":"EQUALS", "value":"rs12460890"}])
print(samples)
[{u'Ref_Counts': u'130', u'href': u'/api/v1/SampleAse/1418785', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418785/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418785/SNP_ID'}, u'Alt_Counts': u'121', u'ID': u'1418785', u'Chromosome': u'19'}, {u'Ref_Counts': u'4142', u'href': u'/api/v1/SampleAse/1418786', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418786/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418786/SNP_ID'}, u'Alt_Counts': u'4791', u'ID': u'1418786', u'Chromosome': u'19'}, {u'Ref_Counts': u'19', u'href': u'/api/v1/SampleAse/1418787', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418787/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418787/SNP_ID'}, u'Alt_Counts': u'28', u'ID': u'1418787', u'Chromosome': u'19'}, {u'Ref_Counts': u'19', u'href': u'/api/v1/SampleAse/1418788', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418788/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418788/SNP_ID'}, u'Alt_Counts': u'23', u'ID': u'1418788', u'Chromosome': u'19'}, {u'Ref_Counts': u'32', u'href': u'/api/v1/SampleAse/1418789', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418789/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418789/SNP_ID'}, u'Alt_Counts': u'11', u'ID': u'1418789', u'Chromosome': u'19'}, {u'Ref_Counts': u'639', u'href': u'/api/v1/SampleAse/1418790', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418790/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418790/SNP_ID'}, u'Alt_Counts': u'572', u'ID': u'1418790', u'Chromosome': u'19'}, {u'Ref_Counts': u'202', u'href': u'/api/v1/SampleAse/1418791', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418791/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418791/SNP_ID'}, u'Alt_Counts': u'309', u'ID': u'1418791', u'Chromosome': u'19'}, {u'Ref_Counts': u'423', u'href': u'/api/v1/SampleAse/1418792', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418792/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418792/SNP_ID'}, u'Alt_Counts': u'401', u'ID': u'1418792', u'Chromosome': u'19'}, {u'Ref_Counts': u'271', u'href': u'/api/v1/SampleAse/1418793', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418793/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418793/SNP_ID'}, u'Alt_Counts': u'234', u'ID': u'1418793', u'Chromosome': u'19'}, {u'Ref_Counts': u'806', u'href': u'/api/v1/SampleAse/1418794', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418794/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418794/SNP_ID'}, u'Alt_Counts': u'1081', u'ID': u'1418794', u'Chromosome': u'19'}, {u'Ref_Counts': u'213', u'href': u'/api/v1/SampleAse/1418795', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418795/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418795/SNP_ID'}, u'Alt_Counts': u'201', u'ID': u'1418795', u'Chromosome': u'19'}, {u'Ref_Counts': u'74', u'href': u'/api/v1/SampleAse/1418796', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418796/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418796/SNP_ID'}, u'Alt_Counts': u'96', u'ID': u'1418796', u'Chromosome': u'19'}, {u'Ref_Counts': u'730', u'href': u'/api/v1/SampleAse/1418797', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418797/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418797/SNP_ID'}, u'Alt_Counts': u'655', u'ID': u'1418797', u'Chromosome': u'19'}, {u'Ref_Counts': u'584', u'href': u'/api/v1/SampleAse/1418798', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418798/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418798/SNP_ID'}, u'Alt_Counts': u'699', u'ID': u'1418798', u'Chromosome': u'19'}, {u'Ref_Counts': u'331', u'href': u'/api/v1/SampleAse/1418799', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418799/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418799/SNP_ID'}, u'Alt_Counts': u'391', u'ID': u'1418799', u'Chromosome': u'19'}, {u'Ref_Counts': u'13', u'href': u'/api/v1/SampleAse/1418800', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418800/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418800/SNP_ID'}, u'Alt_Counts': u'14', u'ID': u'1418800', u'Chromosome': u'19'}, {u'Ref_Counts': u'70', u'href': u'/api/v1/SampleAse/1418801', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418801/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418801/SNP_ID'}, u'Alt_Counts': u'101', u'ID': u'1418801', u'Chromosome': u'19'}, {u'Ref_Counts': u'47', u'href': u'/api/v1/SampleAse/1418802', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418802/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418802/SNP_ID'}, u'Alt_Counts': u'35', u'ID': u'1418802', u'Chromosome': u'19'}, {u'Ref_Counts': u'19', u'href': u'/api/v1/SampleAse/1418803', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418803/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418803/SNP_ID'}, u'Alt_Counts': u'28', u'ID': u'1418803', u'Chromosome': u'19'}, {u'Ref_Counts': u'44', u'href': u'/api/v1/SampleAse/1418804', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418804/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418804/SNP_ID'}, u'Alt_Counts': u'47', u'ID': u'1418804', u'Chromosome': u'19'}, {u'Ref_Counts': u'60', u'href': u'/api/v1/SampleAse/1418805', u'SampleIds': {u'href': u'/api/v1/SampleAse/1418805/SampleIds'}, u'Position': 829568, u'SNP_ID': {u'href': u'/api/v1/SampleAse/1418805/SNP_ID'}, u'Alt_Counts': u'55', u'ID': u'1418805', u'Chromosome': u'19'
}]
There they are.
Let's format the expression counts
for sample in samples:
print("{Ref_Counts:5} {Alt_Counts:5}".format(**sample))
130 121
4142 4791
19 28
19 23
32 11
639 572
202 309
423 401
271 234
806 1081
213 201
74 96
730 655
584 699
331 391
13 14
70 101
47 35
19 28
44 47
60 55
Let's plot the expression counts in these samples in a scatter plot.
import matplotlib.pyplot as plt
plt.scatter([sample["Ref_Counts"] for sample in samples], [sample["Alt_Counts"] for sample in samples])
plt.xlim([0, 5000])
plt.ylim([0, 5000])
plt.xlabel("Reference Allele")
plt.ylabel("Alternative Allele")
plt.title("Allele-Specific Expression for rs12460890")
And add a line for the non-specific expression.
plt.plot([0, 5000], [0, 5000])
plt.show()