The R API is a script that allows access to your MOLGENIS data via the R-Project statistical package. It is available on every MOLGENIS on the URL http://molgenis.mydomain.example/molgenis.R.
NOTE: The MOLGENIS R-api client supports up to R-version 3.2.x
Example
As an example, let's create a plot for publicly available ASE data available on https://molgenis56.target.rug.nl/. For a description of the data, take a look at http://molgenis.org/ase.
Start up the R environment.
In the shell type:
library('RCurl')
eval(expr = parse(text = getURL("https://molgenis56.target.rug.nl/molgenis.R")))
This loads the R API from the molgenis56 server. If you take a look in your workspace by typing
ls()
you should see that a couple functions have been added for you to use:
[1] "molgenis.add" "molgenis.addAll" "molgenis.addList" "molgenis.delete" "molgenis.env"
[6] "molgenis.get" "molgenis.getAttributeMetaData" "molgenis.getEntityMetaData" "molgenis.login" "molgenis.logout"
[11] "molgenis.update"
Let's load some data from the server using molgenis.get:
molgenis.get("ASE")
This retrieves the top 1000 rows from the ASE entity.
P_Value Samples SNP_ID Chr Pos Genes
1 0.000000000000000020650473933963698652198164782417962682333833636491755847419682368126814253628253936767578125000000000000000000000000000000000000000000000000000000000000000000000 145 rs9901673 17 7484101 ENSG00000129226,ENSG00000264772
2 0.000000000000000008781097353981130661746700850633192724259771276345502150073585312384238932281732559204101562500000000000000000000000000000000000000000000000000000000000000000000 359 rs2597775 4 17503382 ENSG00000151552
3 0.000000000000000001491745894983400057481059632909089858257546335023040629669255352496293198782950639724731445312500000000000000000000000000000000000000000000000000000000000000000 301 rs3216 11 214421 ENSG00000177963
[...]
1000 0.000132500824069775005771554265976419628714211285114288330078125000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000000 47 rs1056019 12 41337435 ENSG00000018236
Let's retrieve a specific SNP from the ASE entity:
molgenis.get("ASE", q="SNP_ID==rs12460890")
Fraction_alternative_allele Likelihood_ratio_test_D Alternative_allele Reference_allele P_Value Samples SNP_ID Chr Pos Genes
1 0.527 56.02079 TRUE C 0.00000000000007170854 21 rs12460890 19 829568 ENSG00000172232
This SNP has a mild but significant allele-specific expression, based on expression counts in 21 samples.
Let's retrieve the samples for this SNP:
samples <- molgenis.get("SampleAse", q="SNP_ID==rs12460890")
print(samples)
SNP_ID SampleIds Ref_Counts Alt_Counts Chromosome Position ID
1 rs12460890 ERS194242 130 121 19 829568 1418785
2 rs12460890 ERS326942 4142 4791 19 829568 1418786
3 rs12460890 ERS327006 19 28 19 829568 1418787
4 rs12460890 SRS353551 19 23 19 829568 1418788
5 rs12460890 SRS271084 32 11 19 829568 1418789
6 rs12460890 SRS375020 639 572 19 829568 1418790
7 rs12460890 SRS375024 202 309 19 829568 1418791
8 rs12460890 SRS375022 423 401 19 829568 1418792
9 rs12460890 SRS375030 271 234 19 829568 1418793
10 rs12460890 SRS375026 806 1081 19 829568 1418794
11 rs12460890 SRS375027 213 201 19 829568 1418795
12 rs12460890 SRS376459 74 96 19 829568 1418796
13 rs12460890 SRS375032 730 655 19 829568 1418797
14 rs12460890 SRS376461 584 699 19 829568 1418798
15 rs12460890 SRS376464 331 391 19 829568 1418799
16 rs12460890 SRS376469 13 14 19 829568 1418800
17 rs12460890 SRS376467 70 101 19 829568 1418801
18 rs12460890 SRS376468 47 35 19 829568 1418802
19 rs12460890 SRS418748 19 28 19 829568 1418803
20 rs12460890 SRS418754 44 47 19 829568 1418804
21 rs12460890 SRS418755 60 55 19 829568 1418805
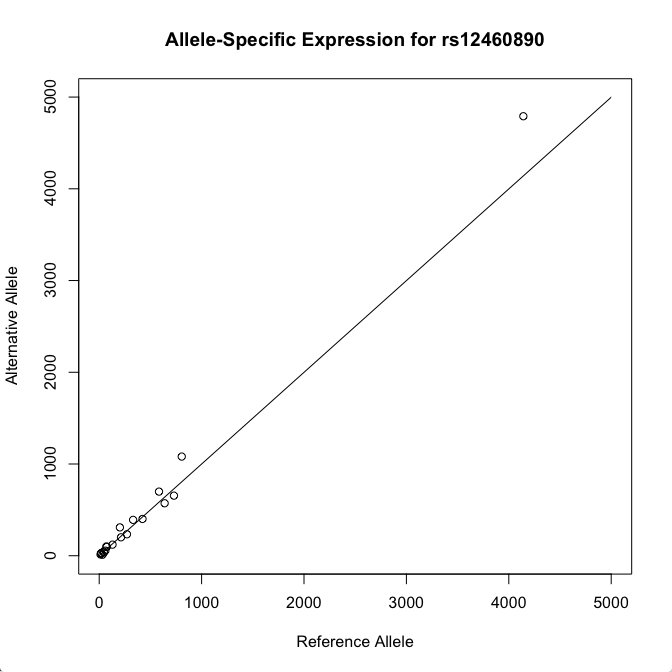
There they are. Let's plot the expression counts in these samples in a scatter plot.
plot(samples$Ref_Counts, samples$Alt_Counts, xlim = c(0, 5000), ylim = c(0, 5000), xlab='Reference Allele', ylab='Alternative Allele', main = 'Allele-Specific Expression for rs12460890')
And add a line for the non-specific expression.
lines(c(0,5000), c(0, 5000))
Query format
The query must be in fiql/rsql format.
Non-anonymous access
The ASE data from the previous example is publicly available. To access private data, you can log in using
molgenis.login("your username", "your password")
This will create a molgenis token on the server and set it in the molgenis.token variable in your R workspace.
When you're done, you can log out using
molgenis.logout()
Retrieving more rows
By default, molgenis.get will retrieve up to 1000 rows only.
If you need more rows, you can request up to 10000 rows by adding the num parameter:
molgenis.get("ASE", num=2000)
will retrieve the top 2000 rows from the ASE entity.
Pagination
You can retrieve the data page-by-page.
molgenis.get("ASE", num=5)
will retrieve a first page of 5 rows and
molgenis.get("ASE", num=5, start=5)
will retrieve the second page of 5 rows.
Cross server scripting
By default, the MOLGENIS R API retrieves its data from the server you retrieved it from, so if you want to retrieve data from a different server, simply source the molgenis.R from that server.
But if you want to combine data from multiple server, you can specify a different REST API URL to use by setting molgenis.api.url in the molgenis.env environment. For example:
local({
molgenis.api.url <- "https://molgenis.mydomain.example:443/api/v1/"
}, env = molgenis.env)
or
local({
molgenis.api.url <- "http://molgenis.mydomain.example:8080/api/v1/"
}, env = molgenis.env)